In
this post Dr. Christian Yates summarises his recent paper “Discrete and continuous models for tissue growth
and shrinkage”, on modelling tissue growth and shrinkage using mathematical models that explicitly incorporate randomness in the tissue deformation process.
The mathematical theme which underlies my research is the development of methodologies for modelling complex biological systems in which randomness (often referred to as stochasticity) plays an important role. In particular, I am interested in modelling complex processes in which the incorporation of noise can produce mean behaviour that differs significantly from the behaviour of a corresponding deterministic model. I am also interested in modelling systems for which, because of their inherent dependence on noise, there is no deterministic counterpart.
The
incorporation of domain growth into stochastic models of biological processes
is of increasing interest to mathematical modellers and biologists alike. In
many situations, especially in developmental biology, the growth of the
underlying tissue domain plays an important role in the redistribution of
particles (be they cells or molecules) which may move and react atop the
domain. Although such processes have largely been modelled using deterministic
(non-random), continuum models, there is an increasing appetite for
individual-based stochastic (random) models, which can capture the fine detail
of the biological movement processes that are being elucidated by modern
experimental techniques, and can also incorporate the inherent stochasticity of
such systems.
I recently had a paper published in the Journal of Theoretical Biology on this subject. In this paper I study a simple stochastic
model of domain growth/shrinkage. From a basic version of this model, Hywood et al. were able to derive a Fokker-Plank equation (FPE) (in this case, an
advection-diffusion partial differential equation on a growing domain), which
describes the evolution of the probability density of some tracer particles on
the domain. My paper extends their work so that a variety of different domain
growth mechanisms can be incorporated. I demonstrate a good agreement between
the mean tracer density and the solution of the FPE in each case. In addition
I incorporate domain shrinkage (via element death) into my individual-level
model and demonstrate that I am also able to derive coefficients for the FPE in
this case. For situations in which the drift and diffusion coefficients
are not readily available, I introduce a numerical coefficient estimation approach.
The basic
individual-based model I use to represent tissue growth/shrinkage is a one-dimensional
domain of made up initially of contiguous elements each of length Δ. I incorporate
growth and shrinkage into this individual-level model by allowing these
elements to undergo ‘proliferation events’ and ‘death events’, which are
analogous to biological cell division and cell death. In order to better understand
the dynamics of the domain growth/shrinkage process, we can place tracer
particles on top of a subset of the domain elements. We say that these domain
elements are ‘marked’. The movement of the domain elements and the tracer
particles resulting from a growth or death event are summarised in Figures 1
and 2.
 |
| (a) |
 |
| (b) |
Figure 1: Examples of growth and division events. Domain elements
are white boxes and tracer particles are represented by smaller red boxes atop
particular ‘marked’ elements. In each subfigure the top configuration shows a domain
before a growth event and the bottom a domain configuration after a growth
event. (a) An unmarked element is chosen to divide. It does so by pushing
itself and the intervals to its right one element length, Δ. Tracer particles move with the
elements and a new element (hatched) is inserted in the empty space. (b) A
marked element is selected to divide. It undergoes the
same movement procedure as for the unmarked element taking its tracer particle with
it. Again a new element (hatched) is inserted in the vacant space.
 |
| (a) |
 |
| (b) |
 |
| (c) |
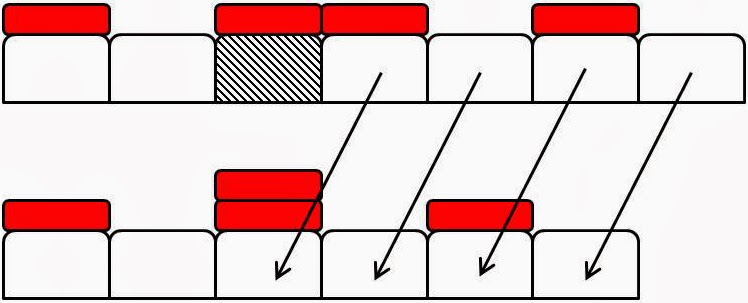
Figure 2: Examples of element
death events. Domain elements are white boxes and tracer particles are
represented by smaller red boxes atop particular ‘marked’ elements. In each
subfigure the top configuration
shows a domain before a death event and the bottom a domain configuration after
a death event. (a) An unmarked element (hatched) is chosen to die. It is
removed from the domain and intervals to its right move leftwards by one
element length, Δ, to fill the space. Tracer
particles move with their elements. (b) A marked element (hatched) is chosen to
die. It is removed from the domain. However its tracer particle remains in place. The elements to the right of the dead
element move to the left one element length, Δ, and a previously
unmarked element becomes marked. (c) A
marked element (hatched) dies and is removed. Its tracer particle remains where
it is and causes the already marked element that was immediately to the right
of the dead element to become doubly marked as it moves into the vacant space.
There is no limit to how many tracer particles an element can accrue.
Deriving the continuum model
By changing the rate at which domain elements grow or die we can incorporate a variety of
different types of domain growth/shrinkage. For each of these different types
of tissue re-arrangement, using the first two infinitesimal moments (specifically the infinitesimal mean, μ, and the infinitesimal
variance, ■) of the domain growth process
we can derive a continuum representation of the density of the tracer particles,
C(x,t), on the domain:
These infinitesimal moments can be generated by considering moment generating
functions of the corresponding birth processes.
Our generalised method allows for the derivation of the
corresponding continuum model for a range of growth rates in the individual-based model: (i) Exponential growth, used to model elongation
of the developing intestinal tract of the quail embryo, growth of sections of
the embryos of the alligator Alligator
mississippiensis, the
initiation and positioning of teeth primordia in the same alligator species and
the early stages of unconstrained cancerous tumour growth); (ii) Linear growth, used to model the early
development of some fish, seeds and body sections of reptile embryos; (iii) Generalised logistic growth, used to model distance
from the dorsal neural tube midline to the distal tip of the lateral mesoderm
in chick embryos (a relevant cell migratory pathway) and the increase in mass
of reptile and bird embryos; (iv) Gompertzian growth, used to model organ
growth, tumour growth and alligator teeth patterning. Comparisons of the
individual-based models and their continuum counterparts for three different growth rates are given in Figure3.
 |
| (b) |
 |
| (c) |
Figure 3: A comparison, at different times, of the expected
occupancy of tracer particles on (a) an exponentially growing domain (b) a linearly
growing domain and (c) a domain growing in according to generalised logistic
growth. The red curves represent the solution of the Fokker-Planck equation and the (noisy) black
curve represents the expected density of tracer particles averaged over 10,000 realisations
of the individual based model. In all three cases the initial
number of domain elements is 60, each of length Δ= ½, with tracer particles initially between 15 ≤ x ≤ 20. The curves are plotted at t = 15, 30, 45, 60.
Domain Shrinkage
The modelling of domain shrinkage
is important for two reasons: (i) the explicit
representation of domain shrinkage is often necessary for a range of
application areas including wound healing, for example, (ii) the
incorporation of element death is important in situations where domain elements
may proliferate and die even if the net growth rate is positive. The second
point is more subtle: it might be argued that domain growth in which element
death is possible but the growth rate, b(t), outweighs the death rate, d(t), can be modelled using a purely
growing domain with a reduced positive growth rate, λ(t) = b(t) − d(t). However, this argument is
incorrect since, although the mean growth rate may be estimated correctly, the
second and higher order moments of the process will be incorrect (c.f. Figure 3 (a) and Figure 4 (a). Both
model exponential domain growth with the same net rate, but Figure 3 (a) shows
the results from a purely growing domain where as domain elements of the domain
used to generate Figure 4 (a) were capable of death as well as growth). In
particular, one stark difference is that the domain in which death is
incorporated explicitly will shrink to zero size with a non-zero probability,
whereas there is no possibility of this happening in the purely growing domain
with reduced net growth rate.
In order to illustrate the
importance of domain shrinkage we incorporate elemental death into the model of
domain growth and also consider a pure domain shrinkage model (see Figure 4).
The infinitesimal moments required to populate the Fokker-Planck equation can
be found by generalising the moment generating function approach to birth-death
processes.
 |
| (a) |
 |
| (b) |
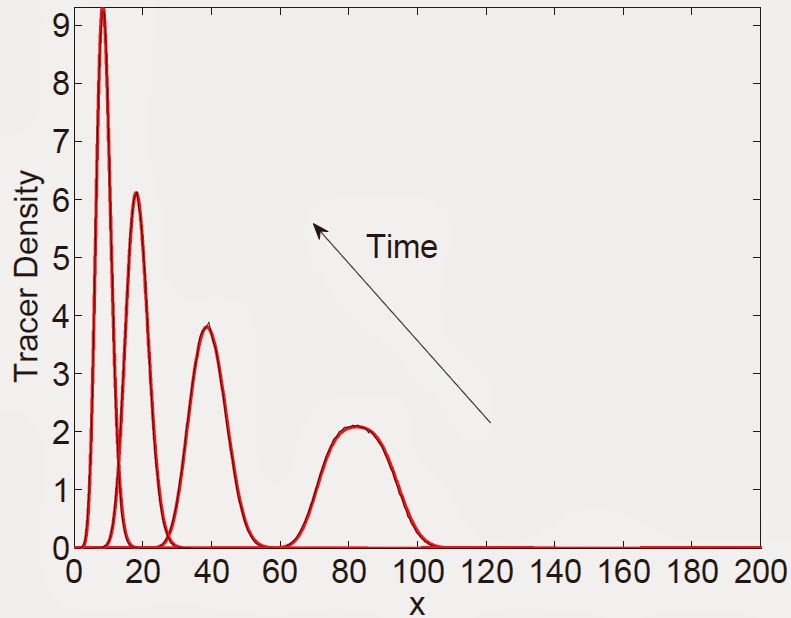
Figure 4: A comparison, at different times, of the expected
occupancy of tracer particles on (a) a domain growing exponentially with constant birth and death
rates, b > d (b) an exponentially shrinking domain with constant death rate, d. Figure descriptions and initial conditions for (a) are as in Fig. 3. For
(b) the initial number of domain elements is 600, each of length Δ= ½, with tracer particles
initially between 150 ≤ x ≤ 200. Note that for (b), the time arrow is in the opposite
direction.
Conclusions
Our generalised method allows
the analytical derivation of the associated drift and diffusion coefficients
for any time-dependent growth rate in the individual-based model. In addition,
we have incorporated the possibility of elemental death into the
individual-based model and derived the coefficients of the corresponding PDE
for these general time-dependent birth and death rates. This approach highlights
that a process in which both elemental birth and death occur cannot simply be
approximated by a birth-only process with the reduced net growth rate since,
although the drift coefficient may be correct, the diffusion coefficient will
not be. Clearly, in situations in which the net birth rate is negative a simple
birth process will not suffice.
For representative examples of
our pure-birth, birth-death and pure-death processes we have carried out
numerical simulations which contrast the expected tracer density in the
individual-level model with the solution of the continuous PDE model and we see
good agreement in each case.
As yet we have considered only the relatively straight-forward case of
uniformly growing domains in which each element is selected to proliferate or die
with equal probability. It is not immediately evident what effect, allowing anisotropic
element proliferation will have on the corresponding drift and diffusion
coefficients. A further challenge will lie in the adaptation of
these methods to multivariate diffusion
processes which will correspond to higher dimensional PDEs. Since tissue growth is an important factor in the
transport of cells across the domain, and often does not occur uniaxially or
uniformly, these extensions will constitute an
important step forward in our ability to model cell migration effectively at both and individual and collective
level.

A discrete system measures variables in discrete or separate points in time. These point in time are usually fixed and are determined based on the system being observed. imp source Discrete models are a way of applying mathematics to biological situations in which the variables are changing over specified time intervals 1.
ReplyDeleteI know there are still a lot of people suffering from different health issues and are therefore looking for solution. I bring you Good news. There is a man called Dr Azuka a herbal practitioner who helped cure me from Genital herpes and shingles, this i had suffered from for the past 2 years and i have spent so much money trying to survive from it. I got my healing by taking the herbal medicine he sent to me to drink for about 14 days . 14 days after completion of the dosage, I went for a medical checkup and I was tested Genital Herpes NEGATIVE. I’m sharing this so that other people can know of this great healer called Dr Azuka because I got to know him through a friend he cured from too. I was made to understand that he can cure several other deadly diseases and infections. Don’t die in ignorance and don’t let that illness take your life. Contact Dr Azuka through his email (dr.azukasolutionhome@gmail.com) or whats app +44 7520 636249
ReplyDeleteHello everyone i am happy to say that i was cure from HIV/AIDS by Dr Padman with his herbal medications after i tested positive july 2016 and ever since then i have been on medications to extend my living on earth. One faithful day i was searching on line if i could get some tips on how to cure my illness and i saw a comment on how Dr Padman cure HIV with herbs,i was surprise and i contact him on the email they provided on the comment and i explain my problem to him and he also prepare the herbs and send to me which i use for one week as i was instructed by him and after 2weeks i went to hospital for checkup and i tested Negative and i am fully cured from the disease and i am so greatful to Dr Padman.You can also reach Dr Padman on his padmanlovespell@yahoo.com , https://twitter.com/padman_dr. https://padmanspell.com WhatsApp +19492293867
ReplyDeleteHOW I WAS ABLE TO FLUSH OUT THE HERPES VIRUS WITH THE HELP OF DR. KHAM NATURAL PRODUCT, HIS EMAIL dr.khamcaregiver@gmail.com. I was diagnosed with Herpes a year ago. It’s the worst thing that’s ever happened to me and I feel like my life is over. Living with constant pain is something no one should have to go through, Herpes has also had a huge impact on my sleep . Since being diagnosed I’ve fallen so far into depression I see no way out because i have been to different hospital. I’m taking a high dose of antidepressants, as well as sedatives and sleeping-pills. I don’t know how I’ll ever accept that this is my life now…I keep asking why me because know changes? But I guess reading your testimony really made me feel happy because i believe what i had from people out there who you also help to cure the herpes virus, and i believe so much with your herbal medicine i am going to be out of this bad situation like others you also help before, what we i have done if not for the herbal medicine that i requested for that was able to flush out the bad virus that was making me go crazy. people out there how will i thank this man called DR. KHAM, i will advice you to email him direct if you have disease like herpes etc. dr.khamcaregiver@gmail.com or whatsapp him +2348159922297
ReplyDeleteI was diagnosed as HEPATITIS B carrier in 2013 with fibrosis of the
ReplyDeleteliver already present. I started on antiviral medications which
reduced the viral load initially. After a couple of years the virus
became resistant. I started on HEPATITIS B Herbal treatment from
ULTIMATE LIFE CLINIC (www.ultimatelifeclinic.com) in March, 2020. Their
treatment totally reversed the virus. I did another blood test after
the 6 months long treatment and tested negative to the virus. Amazing
treatment! This treatment is a breakthrough for all HBV carriers.
I was diagnosed with Idiopathic Pulmonary Fibrosis (IPF) four years ago. For over two years, I relied on prescription medications and therapies, but unfortunately, the symptoms continued to worsen. My breathing became more laboured, and I experienced increasing fatigue and shortness of breath with even minimal activity.Last year, out of desperation and hope, I decided to try an herbal treatment program from NaturePath Herbal Clinic.Honestly, I was skeptical at first, but within a few months of starting the treatment, I began to notice real changes. My breathing became easier, the tightness in my chest eased, and I felt more energetic and capable in my daily life. Incredibly, I also regained much of my stamina and confidence. It’s been a life-changing experience I feel more like myself again, better than I’ve felt in years.If you or a loved one is struggling with IPF, I truly recommend looking into their natural approach. You can visit their website at www.naturepathherbalclinic.com their Email info@naturepathherbalclinic.com
ReplyDelete